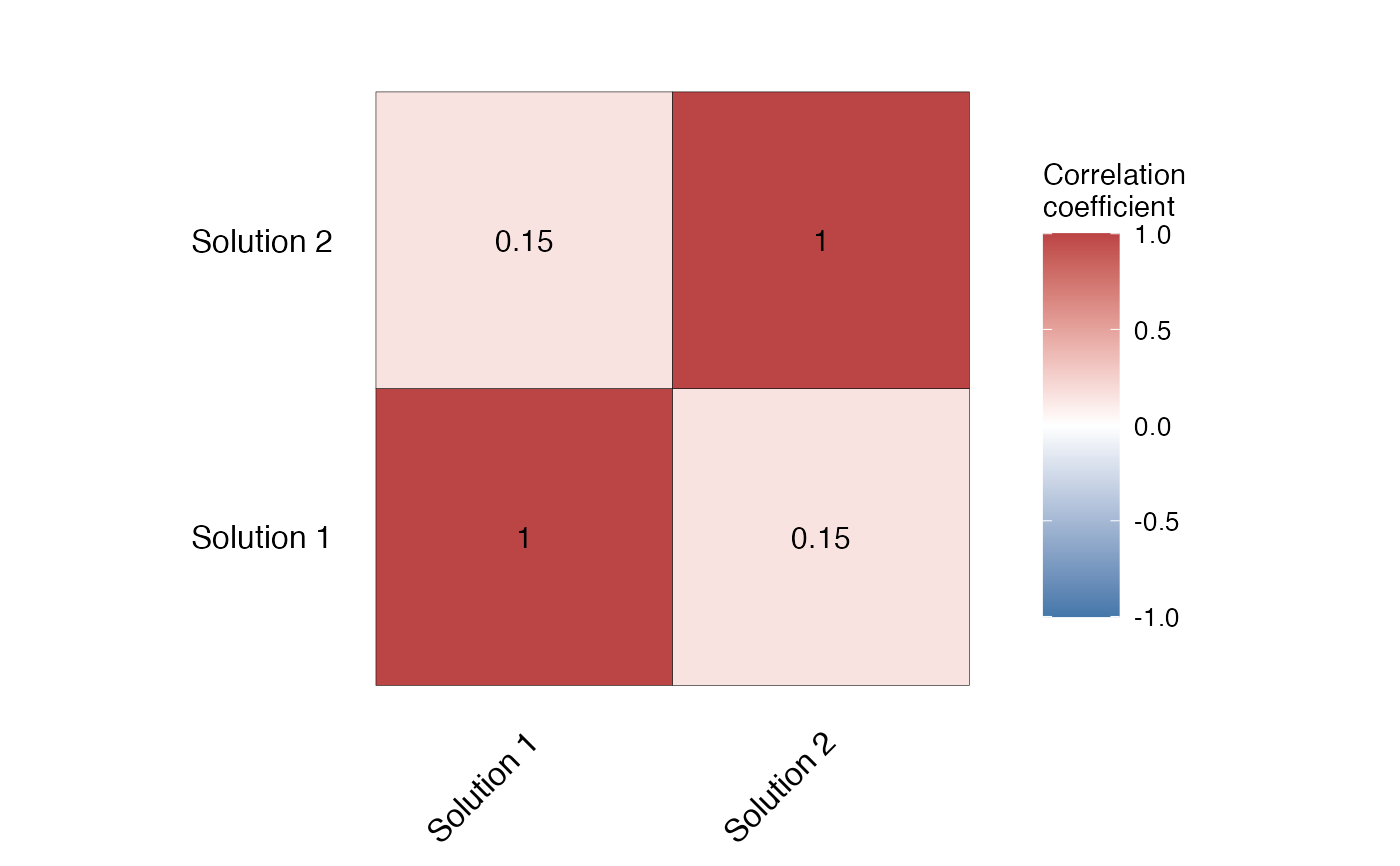
Conservation planning often requires the comparison of the outputs of the solutions of different conservation problems.
One way to compare solutions is by correlating the solutions using Cohen's Kappa. splnr_plot_corrMat() allows to visualize the correlation matrix of the different solutions (for example produced with the spatialplanr function splnr_get_kappaCorrData()).
Usage
splnr_plot_corrMat(
x,
colourGradient = c("#BB4444", "#FFFFFF", "#4477AA"),
legendTitle = "Correlation \ncoefficient",
AxisLabels = NULL,
plotTitle = ""
)Arguments
- x
A correlation matrix of
prioritizrsolutions- colourGradient
A list of three colour values for high positive, no and high negative correlation
- legendTitle
A character value for the title of the legend. Can be empty ("").
- AxisLabels
A list of labels of the solutions to be correlated (Default: NULL). Length needs to match number of correlated solutions.
- plotTitle
A character value for the title of the plot. Can be empty ("").
Examples
# 30 % target for problem/solution 1
dat_problem <- prioritizr::problem(dat_species_bin %>% dplyr::mutate(Cost = runif(n = dim(.)[[1]])),
features = c("Spp1", "Spp2", "Spp3", "Spp4", "Spp5"),
cost_column = "Cost"
) %>%
prioritizr::add_min_set_objective() %>%
prioritizr::add_relative_targets(0.3) %>%
prioritizr::add_binary_decisions() %>%
prioritizr::add_default_solver(verbose = FALSE)
dat_soln <- dat_problem %>%
prioritizr::solve.ConservationProblem()
# 50 % target for problem/solution 2
dat_problem2 <- prioritizr::problem(
dat_species_bin %>%
dplyr::mutate(Cost = runif(n = dim(.)[[1]])),
features = c("Spp1", "Spp2", "Spp3", "Spp4", "Spp5"),
cost_column = "Cost"
) %>%
prioritizr::add_min_set_objective() %>%
prioritizr::add_relative_targets(0.5) %>%
prioritizr::add_binary_decisions() %>%
prioritizr::add_default_solver(verbose = FALSE)
dat_soln2 <- dat_problem2 %>%
prioritizr::solve.ConservationProblem()
CorrMat <- splnr_get_kappaCorrData(list(dat_soln, dat_soln2), name_sol = c("soln1", "soln2"))
#> New names:
#> • `soln1` -> `soln1...1`
#> • `soln1` -> `soln1...2`
#> New names:
#> • `soln2` -> `soln2...1`
#> • `soln2` -> `soln2...2`
#> New names:
#> • `` -> `...1`
#> • `` -> `...2`
(splnr_plot_corrMat(CorrMat, AxisLabels = c("Solution 1", "Solution 2")))
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.